Research Projects
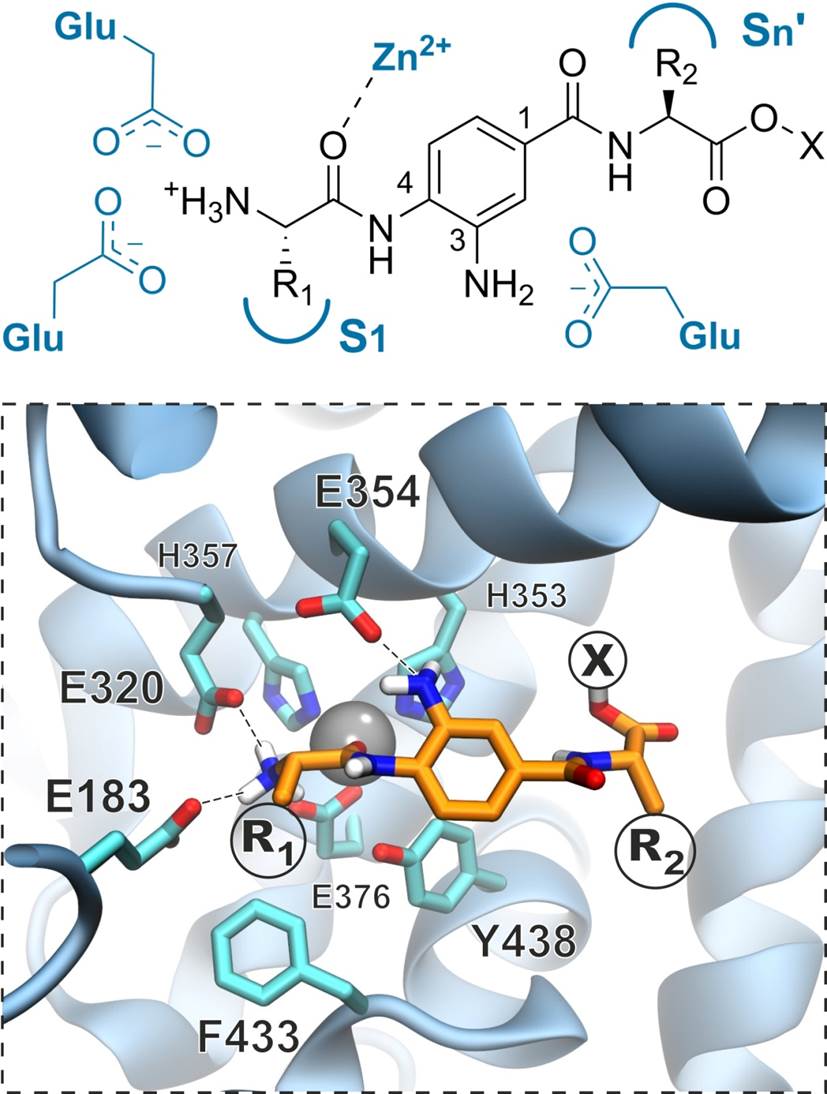
Rational design and synthesis of M1 aminopeptidase inhibitors
We employ structural and biochemical information to design and synthesize selective inhibitors for three members of the oxytocinase subfamily of M1 aminopeptidases, ERAP1, ERAP2 and insulin regulated aminopeptidase (IRAP). These enzymes play important roles in both the adaptive and innate human immune responses and their activity can contribute to the pathogenesis of several major human diseases ranging from viral and parasitic infections to autoimmunity and cancer.
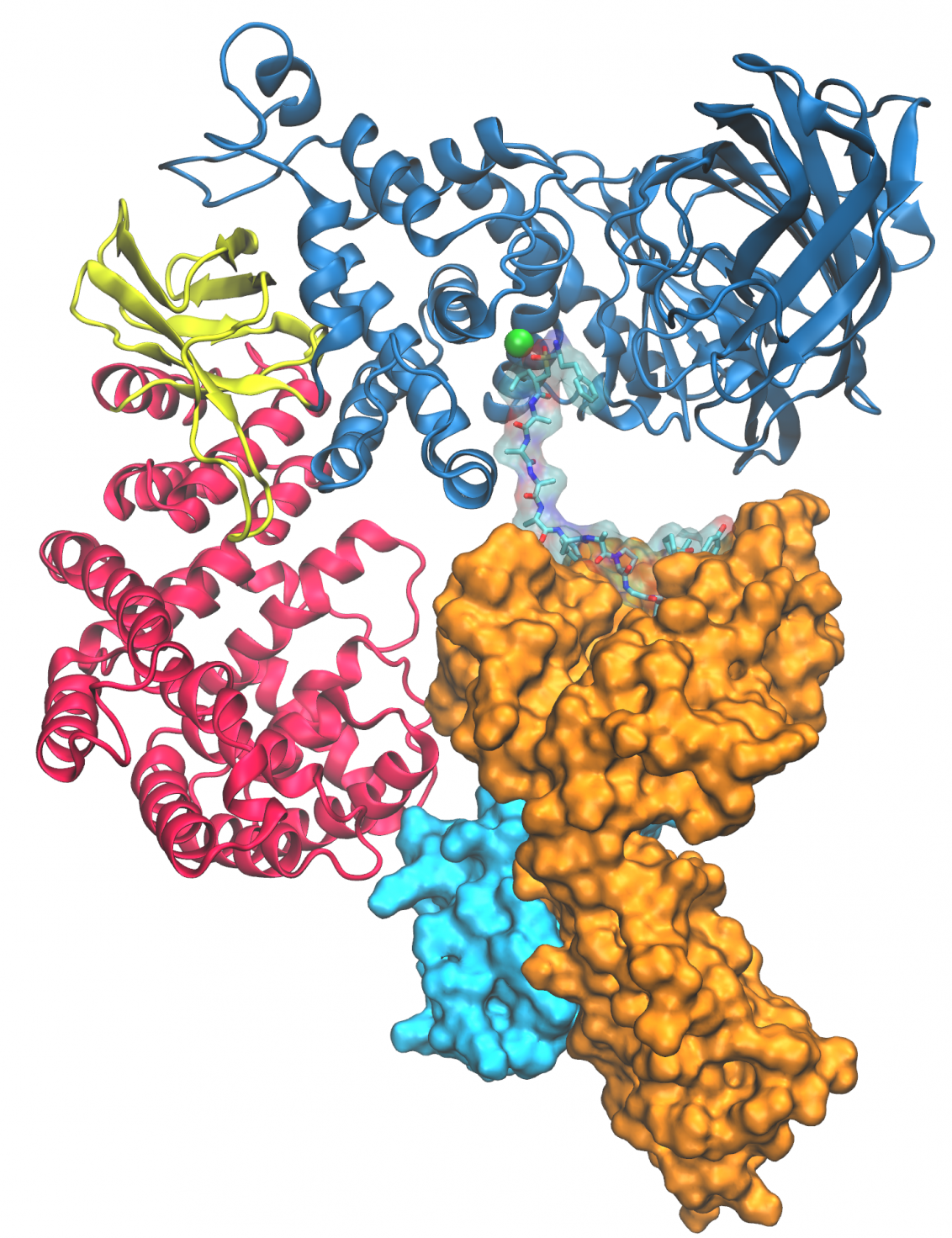
Designed molecules that modulate protein-protein interactions
We aim to elucidate key antigen processing mechanisms of the adaptive immune system at the molecular level using a biochemical investigations and X-ray crystallographic analysis of protein-protein complexes in combination with state-of-the-art computational methods to design peptidomimetic molecules that enhance or disrupt specific protein-protein interactions. Read more about our project "ARIA" by clicking on the picture.
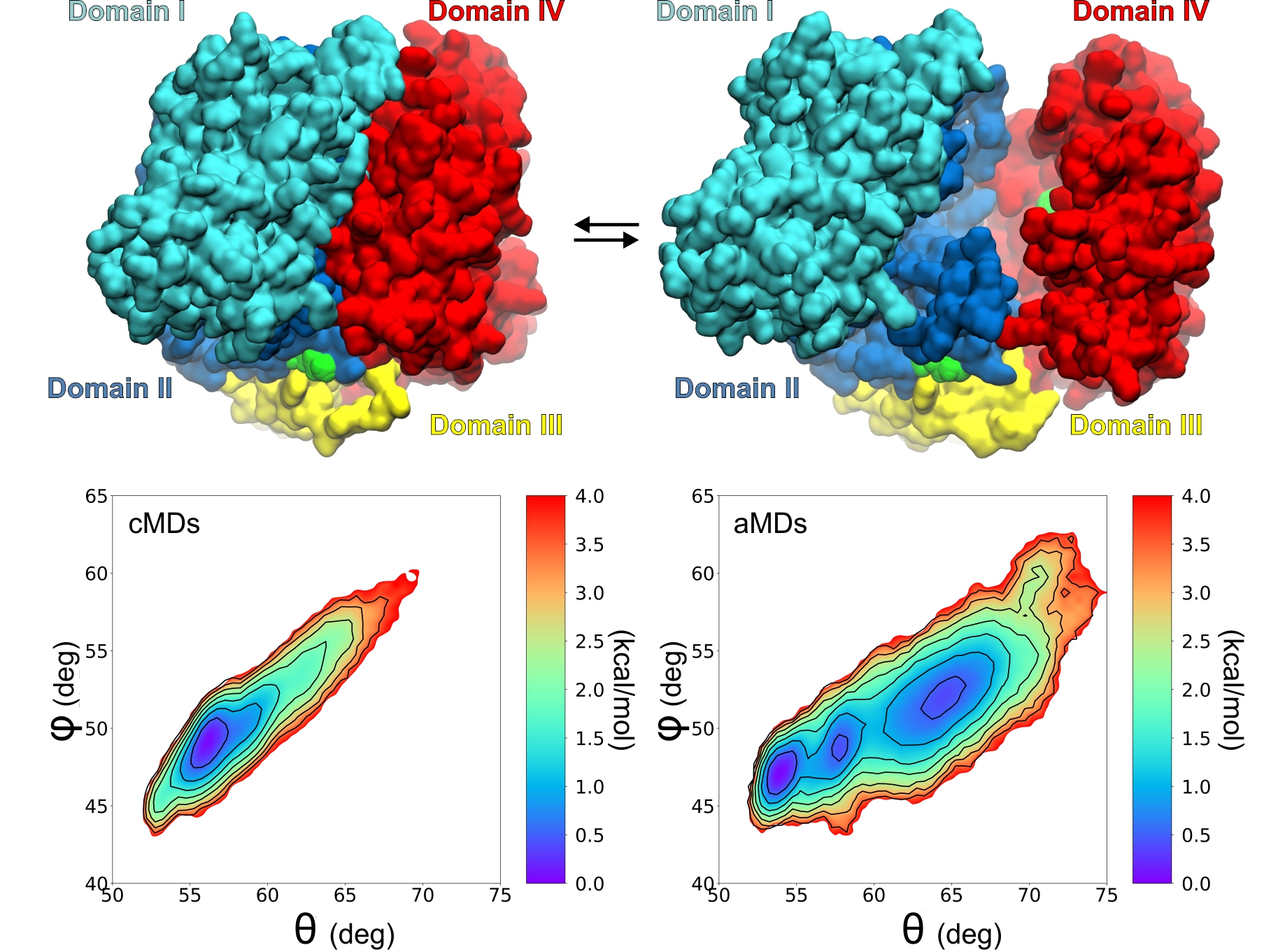
Study of protein dynamics using Molecular Dynamics simulations
We apply state-of-the-art molecular dynamics simulations (MDs) to study the structural dynamics of biomolecules with the aim to understand the link between structure and function. In this context, the dynamics of specific regions of enzymes, or polymorphic positions are investigated with relation to their effect on the enzymatic activity, or interaction with substrates and ligands.
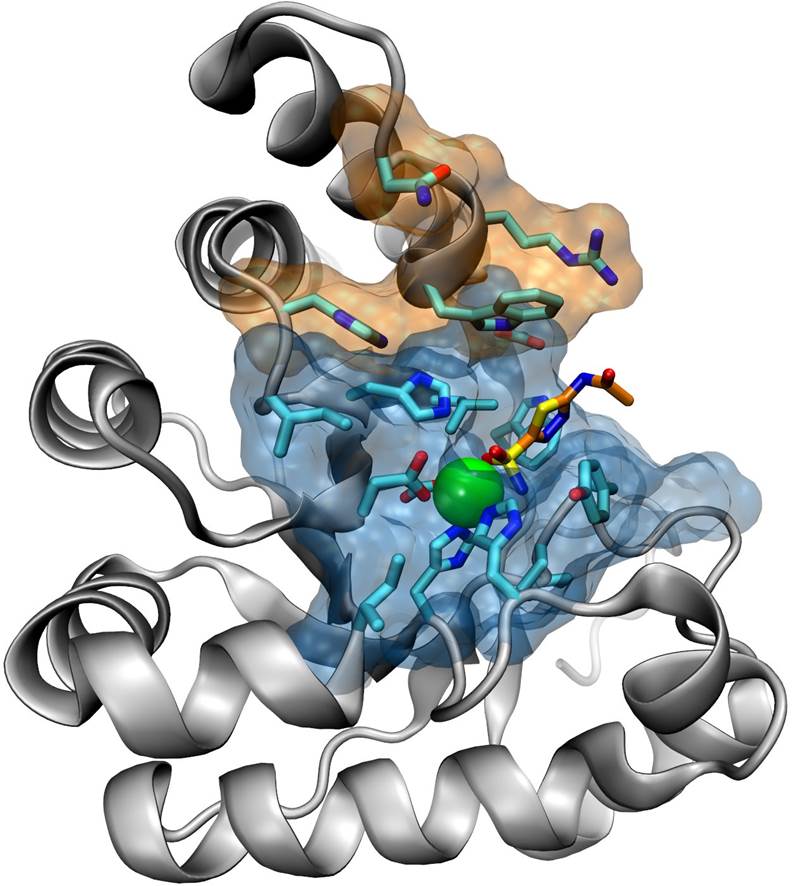
Structure-based discovery of inhibitors for zinc metalloenzymes
We apply state-of-the-art computational methods for the discovery of compounds that target the active site of zinc metalloenzymes, such as the Peptidoglycan N‑Acetylglucosamine Deacetylases. These metalloenzymes are attractive targets for the development of new anti-infective agents against Bacillus cereus, a close relative of the highly virulent Bacillus anthracis
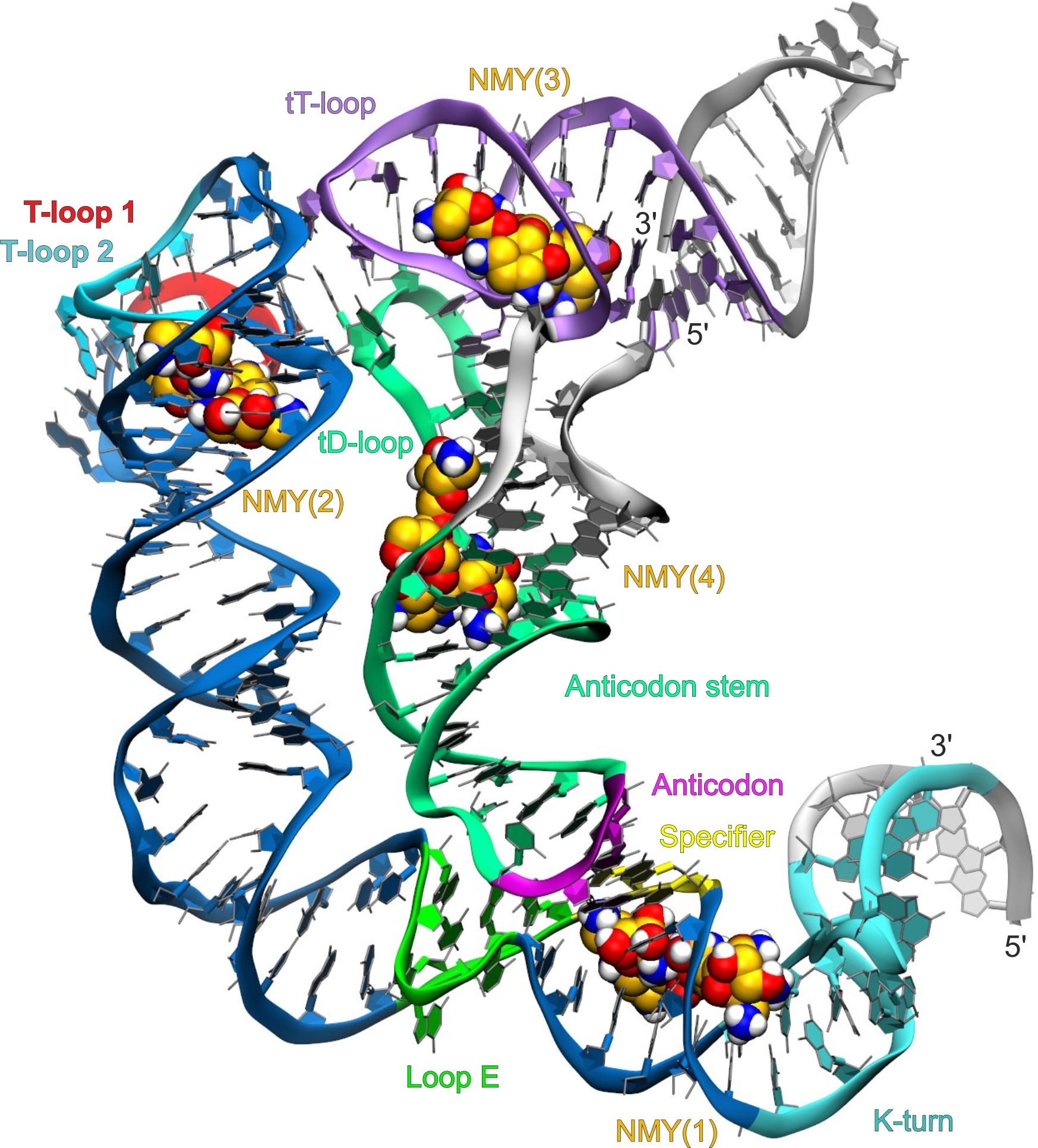
Discovery of RNA-binding molecules as antibacterial agents
T-box riboswitches are among the first discovered transcription attenuators that control mainly transcription of genes related to amino acid transport, metabolism and aminoacylation. Given that T-box riboswitches are found in almost all prominent Gram-positive human pathogens, they represent a promising target for next generation antimicrobial agents. To reach this goal, we apply structure-based virtual screening of large molecular databases to discover novel compounds that stabilize the T-box interaction with its cognite tRNA.
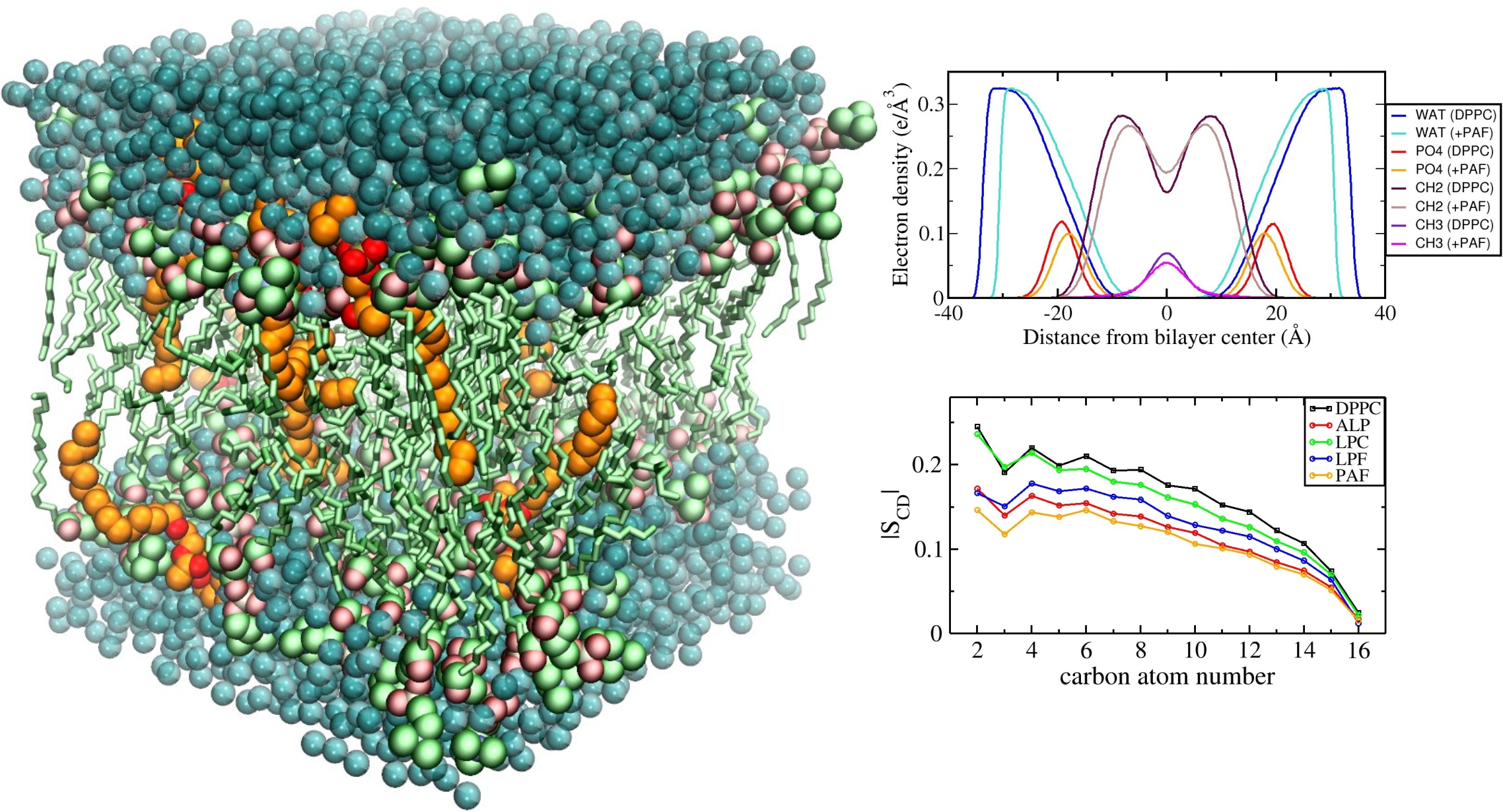
Computational study of membrane properties
We employ computational methods to study the properties of lysophospholipid-containing liposomes, which are intensively studied as drug delivery systems that are stable at normal body temperature, but exhibit fast release of their drug load at slightly elevated temperatures.
Group Leader

Thanos Papakyriakou, PhD
Thanos (Athanasios) is a senior researcher at the Institute of Biosciences & Applications of NCSR "Demokritos" since September 2018. He studied chemistry and obtained his PhD at the National and Kapodistrian University of Athens (2004). He has worked at the University of Florence), at the University of California, San Diego (US) and at the University of Southampton (UK). His research is mainly focused on the design and synthesis of small-molecule inhibitors of enzymes, the structure-based discovery of inhibitors and the computational study of protein structure and dynamics.
Selected Publications
Rational design, synthesis and pharmacological evaluation of a cohort of novel beta-adrenergic receptors ligands enables an assessment of structure-activity relationships
Tricomi, J., Landini, L., Nieddu, V., Cavallaro, U., Baker, J.G., Papakyriakou, A.* Richichi, B.*
European Journal of Medicinal Chemistry 2023, 246, 114961
Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on α-Hydroxy-β-amino Acid Derivatives of Bestatin
Vourloumis, D., Mavridis, I. Athanasoulis, A., Temponeras, I., Koumantou, D., Giastas, P., Mpakali, A., Magrioti, V., Leib, J., van Endert, P., Stratikos, E.*, Papakyriakou, A.*
Journal of Medicinal Chemistry 2022, 65(14):10098-10117
National Collaborators
International Collaborators
thpap@bio.demokritos.gr
Institute of Biosciences & Applications
Building 20, Office Y-31
National Centre for Scientific Research “Demokritos”
Patriarchou Grigoriou and Neapoleos 27
GR-15341 Agia Paraskevi
Athens, Greece
Tel: +30 2106503526
Fax: +30 2106511767